I was asked by Marty (Dr. Matin Blaser, the Director of the Center for Advanced Biotechnology and Medicine, Rutgers University) to write a short note on the recent Cell paper reporting on anti-viral activity of bacterial cells producing multicopy single-stranded DNA (msDNA) encoded from retrons by an Israeli group [Milliman et al., ‘Bacterial Retrons Function Anti- Phage Defense’ Cell (2020) 183: 1-11; Bacterial Retrons Function in Anti-Phage Defense]. This paper was also introduced in the News section of a recent issue of Science (2020) 370: 898-899.
Thirty-six years ago, in 1984, at SUNY Stony Brook, we found a peculiar single-stranded DNA in Myxococcus xanthus, a Gram-negative soil bacterium [Yee et al., Cell (1984) 38: 203-209; Multicopy Single-Stranded DNA Isolated from a Gram-negative Bacterium, Myxococcus xanthus]. Thomas Yee, a graduate student from Caltech was initially working to develop a method to detect minor heterogeneity between two isogenic strains of M. xanthus by digesting their chromosomal DNA by four-base cutting restriction enzymes followed by denaturing, mixing together and renaturing. These were then separated by size using gel electrophoresis on a
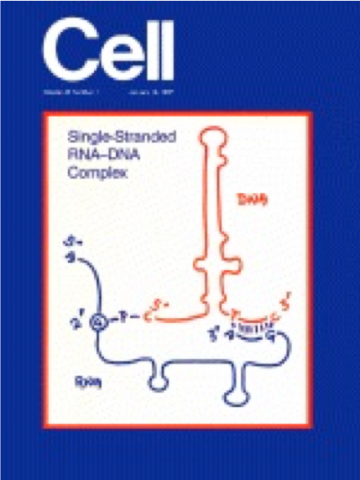
slab gel. Subsequently, the gel strip was cut out, soaked in a buffer containing S1 nuclease, and then placed on a slab gel perpendicular to the first dimension for a second gel electrophoresis. Using this procedure, most DNA molecules are renatured back to their original sizes, which move to the diagonal position in the second gel electrophoresis. However, if there are heterologous fragments, they form a single-stranded tail(s) at the ends of the fragments, which will be removed by S1 nuclease treatment. As a result, these fragments will migrate lower than the diagonal position in the second gel electrophoresis [Yee et al., PNAS (1984) 81: 2723-2727; Two-dimensional S1 nuclease Heteroduplex mapping: Detection of rearrangements in bacterial genomes]. While working on this project, Tom also started to analyze the intact chromosomal DNA of M. xanthus by gel electrophoresis. To our surprise, he found a very stable single-stranded DNA of 162 bases long, existing in high copy number, 500-700 copies per cell, and thus this DNA was termed msDNA for multicopy single-stranded DNA [Yee et al. (1984) Cell 38: 203-209; Multicopy Single-stranded DNA isolated from a Gram-negative bacterium, Myxococcus xanthus]. We then further analyzed this single-stranded DNA and found that it is branched out from the middle of an RNA molecule by a 2’,5’-phosphodiester linkage. These experiments were carried out by two graduate students, T. Furuichi from Japan and A. Dhundale. Two papers of these results were published in Cell. As shown in this figure, a single-stranded DNA is branched out from the middle of RNA using a 2’,5’-phosphodiester linkage, and the 3’ ends of both DNA and RNA form a double-stranded structure (see Figure 1). It looks like an image of a dinosaur; the 5’ end of RNA is the head, two stem-loop structures of RNA are legs in blue, and the secondary structure of DNA in red is a lump on the back of the dinosaur. [Furuichi et al. (1987) Cell 48: 47-53; Branched RNA covalently linked to the 5’ end of a single-stranded DNA in Stigmatella aurantiaca], and [Furuichi et al. (1987) Cell 48: 55-62; Biosynthesis and structure of stable brached RNA covalently linked to the 5’ end of multicopy single-stranded DNA of Stigmatella aurantiaca]. To our delight, the structure of the RNA-DNA complex of msDNA was used for the cover of the Cell issue (Vol. 48) as shown in Figure 1.
We further worked to determine whether a reverse transcriptase is required for the biosynthesis of
msDNA, whose gene was found downstream of the region encoding the precursor RNA for msDNA, and for the first time we showed the existence of reverse transcriptases in bacteria [Lampson et al. (1989) Science 243: 1033-1038. Reverse transcriptase in a clinical strain of Escherichia coli: Production of branched RNA-linked msDNA], [Lamspon et al. (1989) Cell 56: 701-707. Reverse transcriptase with concomitant ribonuclease H activity in the cell-free system of branched RNA-linked msDNA of Myxococcus xanthus], and [Inouye et al. (1989) Cell Reverse transcriptase associated with the biosynthesis of the branched RNA-linked msDNA in Myxococcus xanthus].
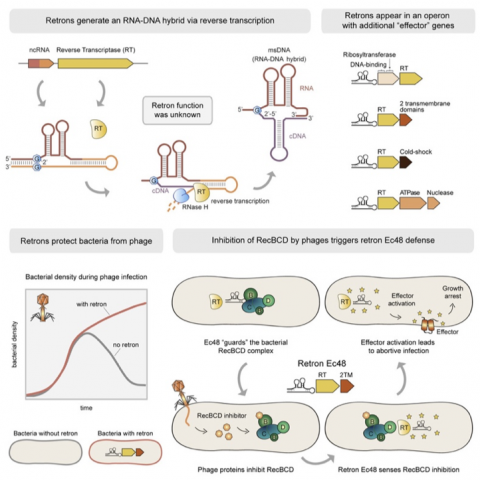
Figure 2 is cited from the Cell paper by Milliman et al., demonstrating how phage infection of cells carrying a retron-producing msDNA results in cell death through the following steps; (1) Phage infection of a cell produces an inhibitor of RecBCD, (2) this inhibitor binds to RecBCD, which is the first line of defense against phages, (3) the RecBAD-inhibitor complex then interacts with msDNA associating with a protein called an effector, and (4) msDNA’s activated effector disrupts the cell membrane to cause cell death to prevent phage proliferation.
Although 36 years sounds like a long time, it is quite amazing that a new phenomenon which happens to be associated with a very old discovery of a factor of unknown function can occur. The authors of the Cell paper pointed out that their finding may lead to a novel therapeutic application to prevent viral infection. Thus, a great lesson is that when we find something of an unknown function, do not stop there, since further characterization of that factor may lead to the opening of a new exciting field in life science.